Physics of Informational Polymers
Formation mechanism of thermally controlled pH gradients

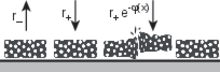
Spatial proton gradients create energy in biological systems and are likely a driving force for prebiotic systems. Due to the fast diffusion of protons, they are however difficult to create as steady state, unless driven by other non-equilibria such as thermal gradients. Here, we quantitatively predict the heat-flux driven formation of pH gradients for the case of a simple acid-base reaction system. To this end, we (i) establish a theoretical framework that describes the spatial interplay of chemical reactions with thermal convection, thermophoresis, and electrostatic forces by a separation of timescales, and (ii) report quantitative measurements in a purpose-built microfluidic device. We show experimentally that the slope of such pH gradients undergoes pronounced amplitude changes in a concentration-dependent manner and can even be inverted. The predictions of the theoretical framework fully reflect these features and establish an understanding of how naturally occurring non-equilibrium environmental conditions can drive pH gradients.
Self-Assembly of Informational Polymers by Templated Ligation
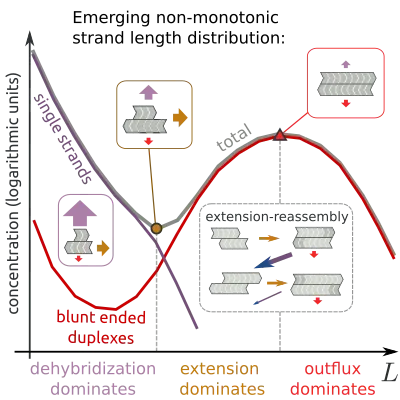
A key aim in origins of life research is to understand how self-replicating evolving systems can spontaneously emerge in non-equilibrium environments. Informational polymers such as RNA play pivotal roles in these scenarios, as information carriers and, according to the “RNA world” hypothesis, also as the first functional molecules catalyzing reactions. However, it is not clear how RNA strands that are long enough to act as functional molecules came into being.
In order to answer this question, the structure that is already induced by the growth of short informational polymers must be understood. Short informational polymers can grow via a process called templated ligation: Two strands bind (hybridize) next to each other on a third strand, the so-called template, which enables the joining (ligation) of the two strands into one.
We studied how this growth process shapes the length distribution of informational polymers. Using stochastic simulations, we find that the process generally leads to a minimum and a maximum in the length distribution. The maximum constitutes a source of informational polymers of a specific length that can be tuned by changing physical parameters. As such, these strands can serve as the building blocks for a structure-forming process on longer length scales. We theoretically characterize the underlying dynamics that shape the length distribution, and confirm the non-monotonous behavior in a proof-of-principle experiment with DNA strands.
Effective dynamics of nucleosome configurations at the yeast PHO5 promoter
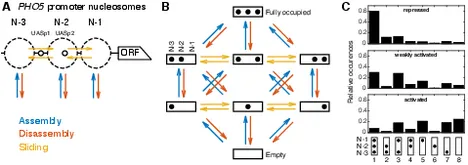
Chromatin dynamics are mediated by remodeling enzymes and play crucial roles in gene regulation, as established in a paradigmatic model, the S. cerevisiae PHO5 promoter. However, effective nucleosome dynamics, i.e. trajectories of promoter nucleosome configurations, remain elusive. Here, we infer such dynamics from the integration of published single-molecule data capturing multi-nucleosome configurations for repressed to fully active PHO5 promoter states with other existing histone turnover and new chromatin accessibility data. We devised and systematically investigated a new class of 'regulated on-off-slide' models simulating global and local nucleosome (dis)assembly and sliding. Only seven of 68145 models agreed well with all data. All seven models involve sliding and the known central role of the N-2 nucleosome, but regulate promoter state transitions by modulating just one assembly rather than disassembly process. This is consistent with but challenges common interpretations of previous observations at the PHO5 promoter and suggests chromatin opening by binding competition.
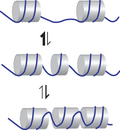
Dynamics of the Buckling Transition in Double-Stranded DNA and RNA
DNA under torsional strain undergoes a buckling transition that is the fundamental step in plectoneme nucleation and supercoil dynamics, which are critical for the processing of genomic information. Despite its importance, quantitative models of the buckling transition, in particular to also explain the surprising two-orders-of-magnitude difference between the buckling times for RNA and DNA revealed by single-molecule tweezers experiments, are currently lacking. Additionally, little is known about the configurations of the DNA during the buckling transition because they are not directly observable experimentally. Here, we use a discrete worm-like chain model and Brownian dynamics to simulate the DNA/RNA buckling transition. Our simulations are in good agreement with experimentally determined parameters of the buckling transition. The simulations show that the buckling time strongly and exponentially depends on the bending stiffness, which accounts for more than half the measured difference between DNA and RNA. Analyzing the microscopic conformations of the chain revealed by our simulations, we find clear evidence for a solenoid-shaped transition state and a curl intermediate. The curl intermediate features a single loop and becomes increasingly populated at low forces. Taken together, the simulations suggest that the worm-like chain model can account semiquantitatively for the buckling dynamics of both DNA and RNA.
The efficiency of driving chemical reactions by a physical non-equilibrium is kinetically controlled
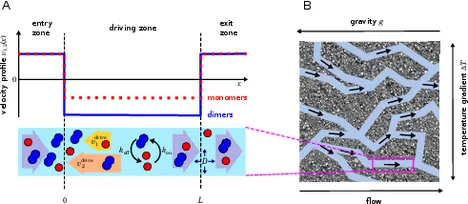
An out-of-equilibrium physical environment can drive chemical reactions into thermodynamically unfavorable regimes. Under prebiotic conditions such a coupling between physical and chemical non-equilibria may have enabled the spontaneous emergence of primitive evolutionary processes. Here, we study the coupling efficiency within a theoretical model that is inspired by recent laboratory experiments, but focuses on generic effects arising whenever reactant and product molecules have different transport coefficients in a flow-through system. In our model, the physical non-equilibrium is represented by a drift–diffusion process, which is a valid coarse-grained description for the interplay between thermophoresis and convection, as well as for many other molecular transport processes. As a simple chemical reaction, we consider a reversible dimerization process, which is coupled to the transport process by different drift velocities for monomers and dimers. Within this minimal model, the coupling efficiency between the non-equilibrium transport process and the chemical reaction can be analyzed in all parameter regimes. The analysis shows that the efficiency depends strongly on the Damköhler number, a parameter that measures the relative timescales associated with the transport and reaction kinetics. Our model and results will be useful for a better understanding of the conditions for which non-equilibrium environments can provide a significant driving force for chemical reactions in a prebiotic setting.
Adsorption-Desorption Kinetics of Soft Particles
Adsorption-desorption processes are ubiquitous in physics, chemistry, and biology. Models usually assume hard particles, but within the realm of soft matter physics the adsorbing particles are compressible. A minimal 1D model reveals that softness fundamentally changes the kinetics: Below the desorption time scale, a logarithmic increase of the particle density replaces the usual Rényi jamming plateau, and the subsequent relaxation to equilibrium can be nonmonotonic and much faster than for hard particles. These effects will impact the kinetics of self-assembly and reaction-diffusion processes.
Replication-guided nucleosome packing and nucleosome breathing expedite the formation of dense arrays
The condensation of eukaryotic chromatin into DNA entails the formation of dense nucleosome arrays. These are frequently disrupted by transcription and replication, such that reassembly is required. The kinetics of this reassembly is of central interest. We investigate scenarios that enable to reach densely packed arrays within biologially reasonable timescale and find that nucleosome breathing, stepwise nucleosome assembly as well as replication guided processes expedite the array assembly. more...
Escalation of polymerization in a thermal gradient
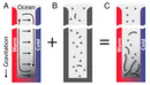
In cells, long DNA and RNA polymers are formed with the help of sophisticated enzymes. However, it is unclear how these information-carrying polymers could have spontaneously formed in the "premordial soup" without enzymes. Here, we demonstrate a mutual positive feedback between the chemical polymerization reaction and a physical non-equilibrium process. This feedback circle leads to a dramatic enhancement of the probability to generate long molecules from dilute solutions of monomers. more...
Toward a unified physical model of nucleosome patterns flanking transcription start sites
The genomes of all eukaryotic organisms are highly packaged into a dynamic structure termed chromatin. On the lowest level of packaging, the structure consists of a "beads-on-a-string" arrangement, where nucleosome "particles" are connected by free DNA "linkers". A large body of recent experimental and theoretical work suggests that this structure can be appropriately described, on a coarse-grained physical level, within the theory of 1D interacting gas systems. In this work, we study which properties are required of such an interacting gas model to yield a "unified" description of nucleosome patterns in different species. more...
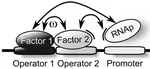
Physical limits on cooperative protein-DNA binding and the kinetics of combinatorial transcription regulation
Much of the complexity observed in gene regulation originates from cooperative protein-DNA binding. While studies of the target search of proteins for their specific binding sites on the DNA have revealed design principles for the quantitative characteristics of protein-DNA interactions, no such principles are known for the cooperative interactions between DNA-binding proteins. We consider a simple theoretical model for two interacting transcription factor (TF) species, searching for and binding to two adjacent target sites hidden in the genomic background. more...
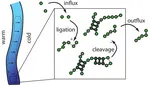
Emergence of information transmission in a prebiotic RNA reactor
A poorly understood step in the transition from a chemical to a biological world is the emergence of self-replicating molecular systems. We study how a precursor for such a replicator might arise in a hydrothermal RNA reactor, which accumulates longer sequences from unbiased monomer influx and random ligation. In the reactor, intra- and intermolecular base pairing locally protects from random cleavage. By analyzing stochastic simulations, we find temporal sequence correlations that constitute a signature of information transmission, weaker but of the same form as in a true replicator. more...
Quantitative test of statistical positioning
Within the last five years, knowledge about nucleosome organization on the genome has grown dramatically. To a large extent this has been achieved by an increasing number of experimental studies determining nucleosome positions at high resolution over entire genomes. Particular attention has been paid to promoter regions, where a canonical pattern has been established: a nucleosome free region with pronounced adjacent oscillations in the nucleosome density. In our study, we tested to what extent this pattern may be quantitatively described by a minimal physical model, a one-dimensional gas of impenetrable particles, commonly referred to as the “Tonks gas”. more...
Further Publications in this Research Area
C. Richert, G. Leveau, D. Pfeffer, B. Altaner, E. Kervio, U. Gerland, F. Welsch (2022)
Enzyme-Free Copying of 12 Bases of RNA with Dinucleotides
Angewandte Chemie International Edition
S. Schink, S. Renner, K. Alim, V. Arnaut, F.C. Simmel, and U. Gerland (2012),
Quantitative Analysis of the Nanopore Translocation Dynamics of Simple Structured Polynucleotides,
Biophysical Journal 102, 85–95.
T. Schötz, R.A. Neher, and U. Gerland (2011),
Target search on a dynamic DNA molecule,
Physical Review E 84, 051911.
N. Geisel and U. Gerland (2011),
Physical limits on cooperative protein-DNA binding and the kinetics of combinatorial transcription regulation,
Biophysical Journal 101, 1569–79.
K. Leu, E. Kervio, B. Obermayer, R. Turk-MacLeod, C. Yuan, J. Luevano, E. Chen, U. Gerland, C. Richert, and I.A. Chen (2013),
Cascade of Reduced Speed and Accuracy After Errors in Enzyme-Free Copying of Nucleic Acid Sequences,
J. Am. Chem. Soc. 135, 354–366.
R. Turk-MacLeod, U. Gerland, and I. Chen (2013)
'Life: the physical underpinnings of replication' in 'Astrochemistry and Astrobiology: Physical Chemistry in Action'
Springer, New York; Eds. I.W.M. Smith, C.S. Cockell, S. Leach; ISBN 978-3-642-31729-3
B. Obermayer, H. Krammer, D. Braun, and U. Gerland (2011),
Emergence of information transmission in a prebiotic RNA reactor,
Physical Review Letters 107, 018101.
K. Leu, B. Obermayer, S. Rajamani, U. Gerland, and I.A. Chen (2011),
The prebiotic evolutionary advantage of transferring genetic information from RNA to DNA,
Nucleic Acids Research 39, 8135-47.
S. Renner, A. Bessonov, U. Gerland, and F.C. Simmel (2010),
Sequence-dependent unfolding kinetics of DNA hairpins studied by nanopore force spectroscopy,
J. Phys.: Condensed Matter 22, 454119.
W. Möbius and U. Gerland (2010),
Quantitative test of the barrier nucleosome model for statistical positioning of nucleosomes up- and downstream of transcription start sites,
PLoS Comp. Biol. 6, e1000891.
J. Megerle, G. Fritz, U. Gerland, K. Jung, J.O. Rädler (2008),
Timing and dynamics of single cell gene expression in the arabinose utilization system,
Biophysical Journal 95, 2103–2115.
R. Neher, W. Möbius, E. Frey, U. Gerland (2007),
Optimal Flexibility for Conformational Transitions in Macromolecules,
Physical Review Letters 99, 178101.
G. Fritz, N. Buchler, T. Hwa and U. Gerland (2007),
Designing sequential transcription logic: a simple genetic circuit for conditional memory,
Systems and Synthetic Biology 1, 89–98.
W. Möbius, R. Neher, and U. Gerland (2006),
Kinetic accessibility of buried DNA sites in nucleosomes,
Physical Review Letters 97, 208102.
R. Bundschuh and U. Gerland (2006),
A puzzle in DNA biophysics,
European Physical Journal E 19, 347–349.
R. Bundschuh and U. Gerland (2006),
Dynamics of intramolecular recognition: Base-pairing in DNA/RNA near and far from equilibrium,
European Physical Journal E 19, 319–329.
M. McCauley, R. Forties, U. Gerland, and R. Bundschuh (2009),
Anomalous scaling in nanopore translocation of structured heteropolymers,
Physical Biology 6, 036006.
W. Möbius, E. Frey, and U. Gerland (2008),
Spontaneous Unknotting of a Polymer Confined in a Nanochannel,
Nano Letters 8, 4518–4522.
R. Neher and U. Gerland (2006),
Intermediate phase in DNA melting,
Physical Review E 73, 030902 (rapid communication).
R. Neher and U. Gerland (2005),
DNA as a programmable viscoelastic nanoelement,
Biophysical Journal 89, 3846–3855.
R. Bundschuh and U. Gerland (2005),
Coupled dynamics of RNA folding and nanopore translocation,
Physical Review Letters 95, 208104.
N.E. Buchler, U. Gerland, and T. Hwa (2005),
Nonlinear protein degradation and the function of genetic circuits,
Proc. Natl. Acad. Sci. USA 102, 9559–9564.
L. Bintu, N.E. Buchler, H. Garcia, U. Gerland, T. Hwa, J. Kondev, T. Kuhlman, and R. Phillips (2005),
Transcriptional regulation by the numbers: applications,
Curr. Opin. Genet. Dev. 15, 125-135.
L. Bintu, N.E. Buchler, H. Garcia, U. Gerland, T. Hwa, J. Kondev & R. Phillips (2005),
Transcriptional regulation by the numbers: models,
Curr. Opin. Genet. Dev. 15, 116-124.
U. Gerland (2005)
Programmierbare Gen-Logik
Bioforum 1-2, 44–45.
R. Neher and U. Gerland (2004),
Dynamics of force-induced DNA slippage,
Physical Review Letters 93, 198102.
U. Gerland, R. Bundschuh, and T. Hwa (2004),
Translocation of structured polynucleotides through nanopores,
Physical Biology 1, 19–26.
N.E. Buchler, U. Gerland, and T. Hwa (2003),
On schemes of combinatorial transcription logic,
Proc. Natl. Acad. Sci. USA 100, 5136–5141.
U. Gerland, R. Bundschuh, and T. Hwa (2003),
Mechanically probing the folding pathway of single RNA molecules,
Biophysical Journal 84, 2831–40.
U. Gerland, J.D. Moroz, and T. Hwa (2002),
Physical constraints and functional characteristics of Transcription Factor-DNA interaction,
Proc. Natl. Acad. Sci. USA 99, 12105–10.
U. Gerland, R. Bundschuh, and T. Hwa (2001),
Force-induced denaturation of RNA,
Biophysical Journal 81, 1324–32.