Physics of Bacteria
Pressure-induced Shape-shifting of Helical Bacteria
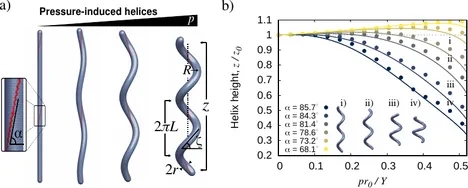
Many bacterial species are helical in form, including the widespread pathogen H. pylori. Motivated by recent experiments on H. pylori showing that cell wall synthesis is not uniform, we investigate the possible formation of helical cell shape induced by elastic heterogeneity. We show, experimentally and theoretically, that helical morphogenesis can be produced by pressurizing an elastic cylindrical vessel with helical reinforced lines. The properties of the pressurized helix are highly dependent on the initial helical angle of the reinforced region. We find that steep angles result in crooked helices with, surprisingly, reduced end-to-end distance upon pressurization. This work helps to explain the possible mechanisms for the generation of helical cell morphologies and may inspire the design of novel pressure-controlled helical actuators
Dynamics of chromosomal target search by a membrane-integrated one-component receptor
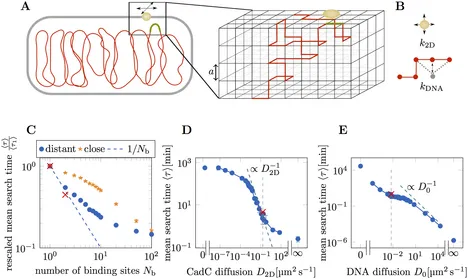
Adaptation to changing environments is vital to bacteria and is enabled by sophisticated signal transduction systems. While signal transduction by two-component systems is well studied, the signal transduction of membrane-integrated one-component systems, where one protein performs both sensing and response regulation, are insufficiently understood. How can a membrane-integrated protein bind to specific sites on the genome to regulate transcription? Here, we study the kinetics of this process, which involves both protein diffusion within the membrane and conformational fluctuations of the genomic DNA. A well-suited model system for this question is CadC, the signaling protein of the E. coli Cad system involved in pH stress response. Fluorescently labeled CadC forms visible spots in single cells upon stable DNA-binding, marking the end of the protein-DNA search process. Moreover, the start of the search is triggered by a medium shift exposing cells to pH stress. We probe the underlying mechanism by varying the number and position of DNA target sites. We combine these experiments with mathematical analysis and kinetic Monte Carlo simulations of lattice models for the search process. Our results suggest that CadC diffusion in the membrane is pivotal for this search, while the DNA target site is just mobile enough to reach the membrane.
Heterogeneous Timing of Gene Induction as a Regulation Strategy
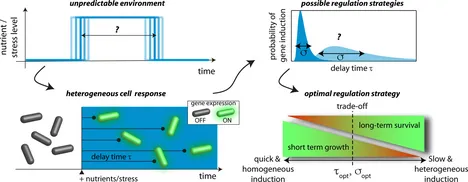
In response to environmental changes, cells often adapt by up-regulating genes to synthesize proteins that generate a benefit in the new environment. Several such cases of gene induction have been reported where the timing was heterogeneous, with some cells responding early and others responding late, although the microbial population was genetically homogeneous and the environment was well mixed. Here, we explore under which conditions heterogeneous timing of gene induction could be advantageous for the population as a whole. We base our study on a mathematical model that accounts for the cost of protein synthesis in terms of resources, which cells must provide immediately, whereas the associated benefit accumulates only slowly over the protein lifetime. Due to this delayed benefit, gene induction can be a risky investment, if resources are scarce and the environment fluctuates rapidly and unpredictably. Unprofitable gene induction then depletes the remaining limiting resource needed for maintenance of cell viability. We show that whenever gene induction is associated with a transient risk but beneficial in the long run, the stochastic timing of gene induction maximizes the reproductive success of a population. In particular, in an environment of stochastic periods of famine and feast, an optimum emerges from a trade-off between short-term growth, favoring rapid and homogeneous responses, and long-term survival, favoring a broadly heterogeneous response. Our analysis suggests that the optimal variability of induction times is just as large as the time required for the amortization of the initial investment into protein synthesis.
Quantifying the benefit of a proteome reserve in fluctuating environments
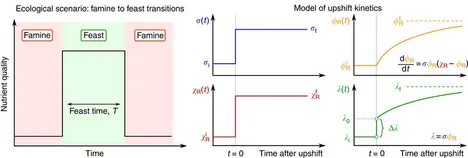
The overexpression of proteins is a major burden for fast-growing bacteria. Paradoxically, recent characterization of the proteome of Escherichia coli found many proteins expressed in excess of what appears to be optimal for exponential growth. Here, we quantitatively investigate the possibility that this overexpression constitutes a strategic reserve kept by starving cells to quickly meet demand upon sudden improvement in growth conditions. For cells exposed to repeated famine-and-feast cycles, we derive a simple relation between the duration of feast and the allocation of the ribosomal protein reserve to maximize the overall gain in biomass during the feast.
Inference of gene regulation functions from dynamic transcriptome data
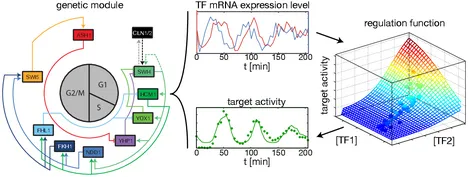
To quantify gene regulation, a function is required that relates transcription factor binding to DNA (input) to the rate of mRNA synthesis from a target gene (output). Such a 'gene regulation function' (GRF) generally cannot be measured because the experimental titration of inputs and simultaneous readout of outputs is difficult. Here we show that GRFs may instead be inferred from natural changes in cellular gene expression, as exemplified for the cell cycle in the yeast S. cerevisiae. We develop this inference approach based on a time series of mRNA synthesis rates from a synchronized population of cells observed over three cell cycles. We first estimate the functional form of how input transcription factors determine mRNA output and then derive GRFs for target genes in the clb2 gene cluster that are expressed during G2/M phase. Systematic analysis of additional GRFs suggests a network architecture that rationalizes transcriptional cell cycle oscillations. We find that a transcription factor network alone can produce oscillations in mRNA expression, but that additional input from cyclin oscillations is required to arrive at the native behaviour of the cell cycle oscillator.
A Dual-Sensing Receptor Confers Robust Cellular Homeostasis
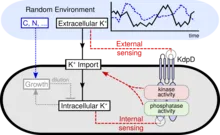
Cells have evolved diverse mechanisms that maintain intracellular homeostasis in fluctuating environments. In bacteria, control is often exerted by bifunctional receptors acting as both kinase and phosphatase to regulate gene expression, a design known to provide robustness against noise. Yet how such antagonistic enzymatic activities are balanced as a function of environmental change remains poorly understood. We find that the bifunctional receptor that regulates K+ uptake in Escherichia coli is a dual sensor, which modulates its autokinase and phosphatase activities in response to both extracellular and intracellular K+ concentration. Using mathematical modeling, we show that dual sensing is a superior strategy for ensuring homeostasis when both the supply of and demand for a limiting resource fluctuate. By engineering standards, this molecular control system displays a strikingly high degree of functional integration, providing a reference for the vast numbers of receptors for which the sensing strategy remains elusive.
A New Way of Sensing: Need-Based Activation of Antibiotic Resistance by a Flux-Sensing Mechanism
Sensing of and responding to environmental changes are of vital importance for microbial cells. Consequently, bacteria have evolved a plethora of signaling systems that usually sense biochemical cues either via direct ligand binding, thereby acting as “concentration sensors,” or by responding to downstream effects on bacterial physiology, such as structural damage to the cell. Here, we describe a novel, alternative signaling mechanism that effectively implements a “flux sensor” to regulate antibiotic resistance. more...
Further Publications
S.A. Westermayer, G. Fritz, J. Gutierrez, J.A. Megerle, M.P.S. Weissl, K. Schnetz, U. Gerland, J. Rädler (2016),
Single-cell characterization of metabolic switching in the sugar phosphotransferase system of Escherichia coli,
Molecular Microbiology (2016) 100(3), 472–485
T. Ramalho, A. Meyer, A. Mückl, K. Kapsner, U. Gerland, F. C. Simmel (2016),
Single Cell Analysis of a Bacterial Sender-Receiver System,
PLoS ONE 11(1): e0145829.
Laure Plener, Nicola Lorenz, Matthias Reiger, Tiago Ramalho, Ulrich Gerland, and Kirsten Jung (2015),
The phosphorylation flow of the Vibrio harveyi quorum sensing cascade determines levels of phenotypic heterogeneity in the population,
J. Bact., 197, 1747-1756.
G. Fritz, J. Megerle, S.A. Westermayer, D. Brick, R. Heermann, K. Jung, J.O. Rädler, and U. Gerland (2014),
Single Cell Kinetics of Phenotypic Switching in the Arabinose Utilization System of E. coli,
PLoS One 9, e89532.
P. Hillenbrand, G. Fritz, and U. Gerland (2013),
Biological signal processing with a genetic toggle switch,
PLoS One 8, e68345
I. Haneburger, G. Fritz, N. Jurkschat, L. Tetsch, A. Eichinger, A. Skerra, U. Gerland, K. Jung (2012),
Deactivation of the E. coli pH Stress Sensor CadC by Cadaverine,
Journal of Molecular Biology, 424, 15–27.
G. Fritz, C. Koller, K. Burdack, L. Tetsch, I. Haneburger, K. Jung, and U. Gerland (2009),
Induction kinetics of a conditional pH stress response system in Escherichia coli,
Journal of Molecular Biology 393, 272–286.
U. Gerland and T. Hwa (2009),
Evolutionary selection between alternative modes of gene regulation,
Proc. Natl. Acad. Sci. USA 106, 8841–8846.
J. Megerle, G. Fritz, U. Gerland, K. Jung, J.O. Rädler (2008),
Timing and dynamics of single cell gene expression in the arabinose utilization system,
Biophysical Journal 95, 2103–2115.