New Publication in Angewandte Chemie International Edition
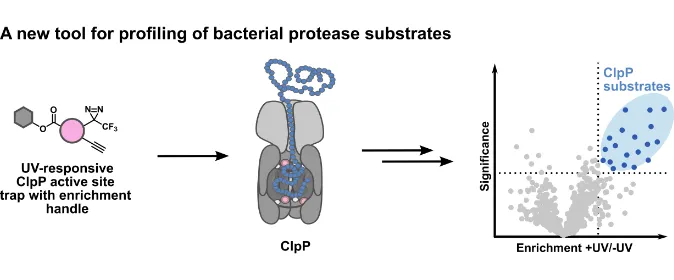
Protein homeostasis in bacteria is regulated by proteases such as the tetradecameric caseinolyticprotease P (ClpP). Although substrates of ClpP have been successfully deciphered in geneticallyengineered cells, methods which directly trap processed proteins within native cells remain elusive.Here, we introduce an in situ trapping strategy which utilizes trifunctional probes that bind to the activesite serine of ClpP and capture adjacent substrates with an attached photocrosslinking moiety. Afterenrichment using an alkyne handle, substrate deconvolution by mass spectrometry (MS) is performed.We show that our two traps bind substoichiometrically to ClpP, retain protease activity, exhibitunprecedented selectivity for Staphylococcus aureus ClpP in living cells and capture numerous knownand novel substrates. The exemplary validation of trapped hits using a targeted proteomics approachconfirmed the fidelity of this technology. In conclusion, we provide a novel chemical platform suited forthe discovery of serine protease substrates beyond genetic engineering.
Gronauer, T. F., Eck, L. K., Ludwig, C., Sieber, S. A. "A Photocrosslinking Probe to Capture the Substrates of Caseinolytic Protease P" Angew. Chem., Int. Ed.
Link: https://doi.org/10.1002/anie.202409220
Copyright: Wiley