Jobs
For B.Sc. and M.Sc. Students
2. Modellierung der Proteinfaltung mit vergröberten Proteinmodellen
Please switch to German language
1. Structure prediction of Protein assemblies based on chemical crosslinking
Most cellular processeand are mediated by large protein complexes (assemblies) and are influenced by transient protein-protein interactions. By using chemical crosslinking it is possible to trapp and analyse these interactions in vitro (in a test tube) but also in vivo (in the cell). Cross linking refers to the covalent connection of two proteins if they come close together (to a distance of less than ~2-3 nm). The crosslink positions on the protein surface can be identified by mass spectrometry. With the knowledge of a sufficient number of such cross links it is in principle possible to determine how the proteins interact and to create a model of the spatial arrangements of the proteins in the molecular assembly or complex. Goal of the master thesis is to design and improve approaches to predict the structure of multi protein complexes based on the structure of the individual proteins and based on the crosslinking data. In the master thesis computational approaches will be used and some knowledge of a programming language like Python is an advantage. If successful the work can have great impact on better understanding biophysical processes in a cell.
2. Describing Proteins as Anisotropic Coarse Densities
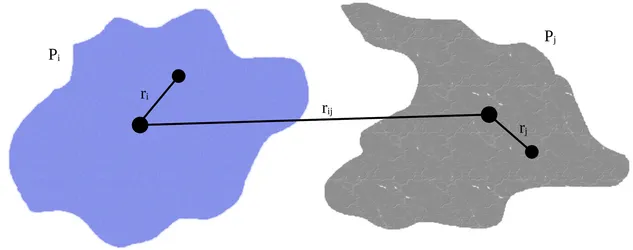
Protein-protein interactions play a central role in life, but understanding these interactions in crowded environments and at larger scales is challenging in simulations. In the last decades, considerable effort has been devoted to the development of coarse-grained force fields that describe proteins and other cellular components with large isotropically interacting beads to simplify the simulation systems and speed up computation. However, the loss of directionality, charge distribution, and deformation due to conformational changes in this coarse description is often problematic in practice and hinders the scientific output of these methods. For this reason, we have recently developed a new approach for coarse-grained simulations that describes proteins as arbitrarily shaped particle densities interacting with each other. This master’s thesis involves the testing and benchmarking of this new method, as well as contributing to the development of simulation and visualization software in Python.