Dr. rer. nat. Martin Kulke
Dr. rer. nat. Martin Kulke
Technische Universität München
Lehrstuhl für Theoretische Biophysik - Molekulardynamik (Prof. Zacharias)
Postadresse
Ernst-Otto-Fischer-Str. 8
85748 Garching b. München
- Tel.: +49 (89) 289 - 13766
- Fax: +49 (89) 289 - 12444
- Raum: 5415.EG.048 Homepage: https://www.groups.ph.tum.de/t38/home/
- martin.kulke@tum.de
Research Interests
- Dynamic properties of protein, lipids and small organic compounds in liquid environments
Biological processes often occur in highly crowded environments, from intracellular condensates and extracellular matrices to complex membrane systems, where proteins, disordered regions, lipids, and small molecules collectively govern structure, dynamics, and function. My research uses molecular dynamics and enhanced sampling to understand how these crowded environments shape biomolecular behavior. I study the assembly and dynamics of stress granules, exploring how intrinsically disordered regions, RNA, and small molecules influence condensate formation, diffusion, and substrate partitioning. In extracellular matrices such as cartilage, I investigate how protein composition, crowding, and aging affect structural stability, mechanical properties, and molecular permeability. Extending these approaches to membranes, I examine how lipid composition, temperature, and membrane proteins regulate dynamics and transport processes. I have also applied simulations to membrane proteins to probe allosteric pathways and structure-function relationships. Through atomistic simulations, my work provides a comprehensive view of how crowded environments influence biomolecular interactions, conformational distributions, and transport.
- Enhanced sampling techniques
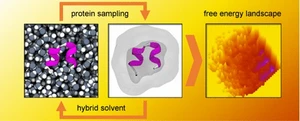
Accurately describing the structural dynamics of large proteins, including intrinsically disordered regions, requires methods that can capture both folded and flexible conformations beyond the reach of current machine learning-based predictors such as AlphaFold and RoseTTAFold. To address this, I developed TIGER2hs (Temperature Intervals with Globally Exchanging Replicas in Hybrid Solvent), a replica-exchange molecular dynamics method that efficiently explores the conformational landscape of proteins and protein-protein interfaces. The approach enables reliable sampling of both folded and disordered ensembles by combining hybrid solvent models with global temperature exchanges. TIGER2hs has been successfully applied to protein folding, conformational ensembles of intrinsically disordered regions, and peptide aggregation, providing detailed insights into how sequence and crowded environments shape conformational distributions.
- Multiscale modeling
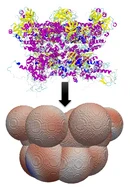
Protein-protein interactions are central to many biological processes, yet capturing their behavior in large crowded cellular environments remains a major challenge for computational simulations. My research focuses on developing an anisotropic supra-coarse-grained simulation framework that bridges atomistic detail and mesoscale modeling. In this approach, proteins are represented as overlapping rigid spheres that diffuse in an implicit solvent governed by overdamped Langevin dynamics. Deterministic and stochastic forces account for molecular shape, hydrodynamic friction, and thermal fluctuations, while interprotein interactions are derived from atomistic potentials to retain key anisotropic and electrostatic features. This framework enables efficient mesoscale simulations of protein transport, assembly, and organization in complex biological environments, helping to connect detailed molecular descriptions with large-scale biological phenomena.
Publications
| Year | Authors, Title, Journal |
| 2025 | Klose, C.J.; Meighen-Berger, K.M.; Kulke, M.; Parr, M.; Steigenberger, B.; Zacharias, M.; Frishman, D.; Feige, M.J. The EMC acts as a chaperone for membrane proteins Nat Commun 2025, 16 (1), 7097 |
| 2025 | Lalaurie, C.J.; Kulke, M.; Geist, N.; Delcea, M.; Dalby, P.A.; McDonnell, T.C.R. The J-shape of β2GPI reveals a cryptic discontinuous epitope across domains I and II Journal of Structural Biology: X 2025, 12, 100135 |
| 2024 | Parson, W.W., Huang, J., Kulke, M., Vermaas J.V., Kramer D.M. Electron transfer in a crystalline cytochrome with four hemes The Journal of Chemical Physics 2024, 160 (6), 065101 |
| 2024 | Kulke, M., Kurtz, E., Boren, D.M., Olson, D.M., Koenig, A.M., Hoffmann-Benning, S., Vermaas, J.V. PLAT domain protein 1 (PLAT1/PLAFP) binds to the Arabidopsis thaliana plasma membrane and inserts a lipid Plant Science 2024, 338, 111900 |
| 2023 | Weraduwage S.M., Whitten, D., Kulke, M., Sahu, A., Vermaas, J.V., Sharkey, T.D. The isoprene-responsive phosphoproteome provides new insights into putative signalling pathways and novel roles of isopren Plant, Cell, & Environment 2023, pce.14776 |
| 2023 | Kulke., M.; Olson, D.M., Huang, J., Kramer, D.M., Vermaas, J.V. Long-Range Electron Transport Rates Depend on Wire Dimensions in Cytochrome Nanowires Small 2023, 19, 2304013 |
2023
| Kulke, M., Weraduwage, S. M., Sharkey, T. D., Vermaas, J. V. Nanoscale Simulation of the Thylakoid Membrane Response to Extreme Temperatures. Plant Cell & Environment 2023, pce.14609 |
| 2023 | Weraduwage, S. M., Sahu, A., Kulke, M., Vermaas, J. V., Sharkey, T. D. Characterization of Promoter Elements of Isoprene‐responsive Genes and the Ability of Isoprene to Bind START Domain Transcription Factors Plant Direct 2023, 7 (2) |
| 2023 | Sarkar, D., Kulke, M., Vermaas, J. V. LongBondEliminator: A Molecular Simulation Tool to Remove Ring Penetrations in Biomolecular Simulation Systems Biomolecules 2023, 13 (1), 107 |
| 2022 | Kulke, M., Vermaas, J. V. Reversible Unwrapping Algorithm for Constant-Pressure Molecular Dynamics Simulations J. Chem. Theory Comput. 2022, 18 (10), 6161–6171 |
| 2022 | Schulig, L., Geist, N., Delcea, M., Link, A., Kulke, M. Fundamental Redesign of the TIGER2hs Kernel to Address Severe Parameter Sensitivity J. Chem. Inf. Model. 2022, 62 (17), 4200–4209 |
| 2022 | Ayala Mariscal, S. M., Pigazzini, M. L., Richter, Y., Özel, M., Grothaus, I. L.,Protze, J., Ziege, K., Kulke, M., ElBediwi, M., Vermaas, J. V., Colombi Ciacchi, L., Köppen, S., Liu, F., Kirstein, J. Identification of a HTT-Specific Binding Motif in DNAJB1 Essential for Suppression and Disaggregation of HTT Nat Commun 2022, 13 (1), 4692 |
| 2021 | Zwicker, P., Geist, N., Göbler, E., Kulke, M., Schmidt, T., Hornschuh, M., Lembke, U., Prinz, C., Delcea, M., Kramer, A., Müller, G. Improved Adsorption of the Antimicrobial Agent Poly (Hexamethylene) Biguanide on Ti-Al-V Alloys by NaOH Treatment and Impact of Mass Coverage and Contamination on Cytocompatibility Coatings 2021, 11 (9), 1118 |
| 2021 | Kulke, M., Nagel, F., Schulig, L., Geist, N., Gabor, M., Mayerle, J., Lerch, M. M., Link, A., Delcea, M. A A Hypothesized Mechanism for Chronic Pancreatitis Caused by the N34S Mutation of Serine Protease Inhibitor Kazal-Type 1 Based on Conformational Studies J. Inflamm. Res. 2021, Volume 14, 2111–2119 |
| 2021 | Schulig, L., Grabarczyk, P., Geist, N., Delin, M., Forkel, H., Kulke, M., Delcea, M., Schmidt, C. A., Link, A. Unveiling the N-Terminal Homodimerization of BCL11B by Hybrid Solvent Replica-Exchange Simulations Int. J. Mol. Sci. 2021, 22 (7), 3650 |
| 2021 | Buchholz, I., McDonnell, T., Nestler, P., Tharad, S., Kulke, M., Radziszewska, A., Ripoll, V. M., Schmidt, F., Hammer, E., Toca-Herrera, J. L., Rahman, A., Delcea, M. Specific Domain V Reduction of Beta-2-Glycoprotein I Induces Protein Flexibility and Alters Pathogenic Antibody Binding Sci. Rep. 2021, 11 (1), 4542 |
| 2020 | Kulke, M., Langel, W. Molecular Dynamics Simulations to the Bidirectional Adhesion Signaling Pathway of Integrin ΑVβ3 Proteins Struct. Funct. Bioinforma. 2020, 88 (5), 679–688 |
| 2019 | Kulke, M., Uhrhan, M., Geist, N., Brüggemann, D., Ohler, B., Langel, W., Köppen, S. Phosphorylation of Fibronectin Influences the Structural Stability of the Predicted Interchain Domain J. Chem. Inf. Model. 2019, 59 (10), 4383–4392 |
| 2019 | Geist, N., Kulke, M., Schulig, L., Link, A., Langel, W. Replica-Based Protein Structure Sampling Methods II: Advanced Hybrid Solvent TIGER2hs J. Phys. Chem. B 2019, 123 (28), 5995–6006 |
| 2019 | Janke, U., Kulke, M., Buchholz, I., Geist, N., Langel, W., Delcea, M. Drug-Induced Activation of Integrin Alpha IIb Beta 3 Leads to Minor Localized Structural Changes PLoS One 2019, 14 (4), e0214969 |
| 2019 | Kulke, M. A Molecular Model for an Adhesion Complex in the Extracellular Matrix of Bone Cells PhD Thesis, University of Greifswald, 2019 |
| 2018 | Kulke, M., Geist, N., Möller, D., Langel, W. Replica-Based Protein Structure Sampling Methods: Compromising between Explicit and Implicit Solvents J. Phys. Chem. B 2018, 122 (29), 7295–7307 |
| 2017 | Kulke, M., Geist, N., Friedrichs, W., Langel, W. Molecular Dynamics Simulations on Networks of Heparin and Collagen Proteins Struct. Funct. Bioinforma. 2017, 85 (6), 1119–1130 |
| 2014 | Aliev, A. E., Kulke, M., Khaneja, H. S., Chudasama, V., Sheppard, T. D., Lanigan, R. M. Motional Timescale Predictions by Molecular Dynamics Simulations: Case Study Using Proline and Hydroxyproline Sidechain Dynamics Proteins Struct. Funct. Bioinforma. 2014, 82 (2), 195–215 |