Welcome to the Biomolecular Dynamics Workgroup
What drives us
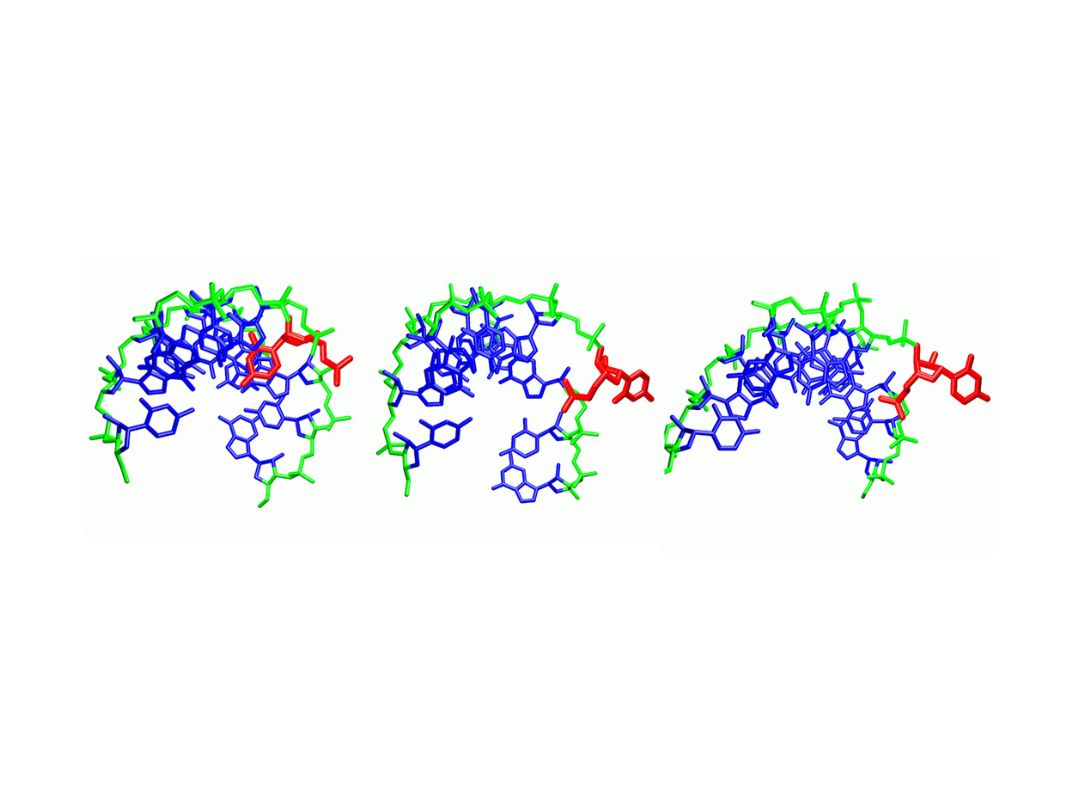
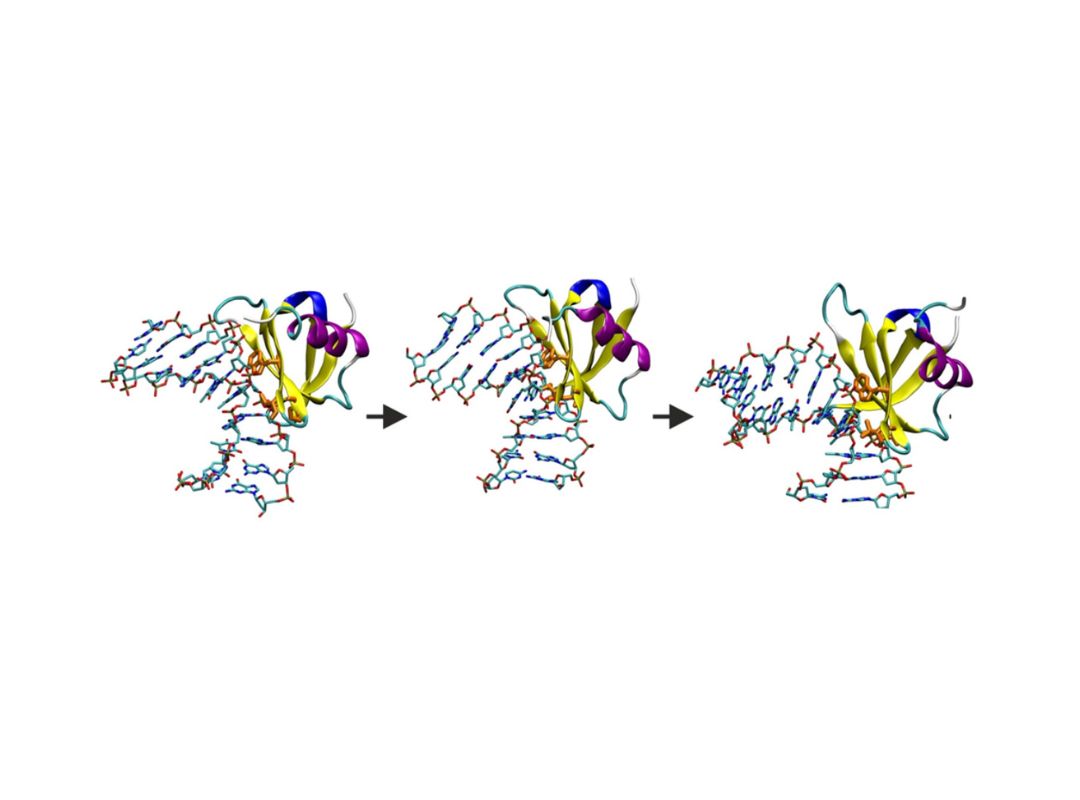
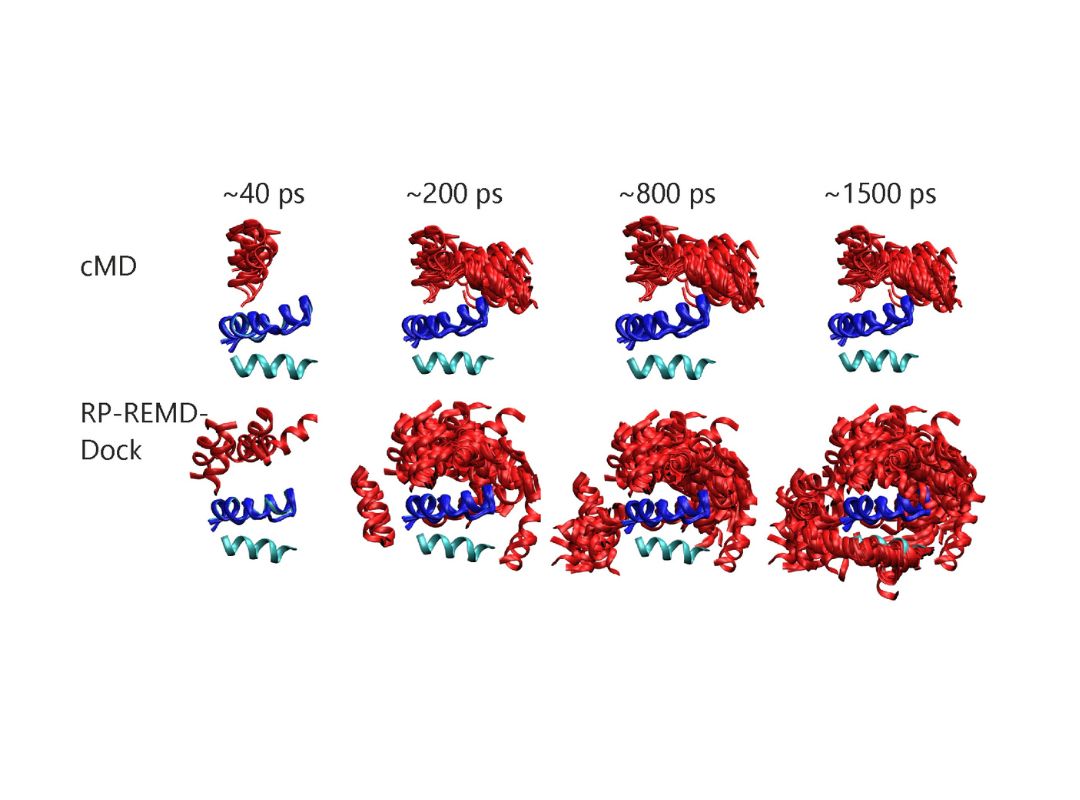
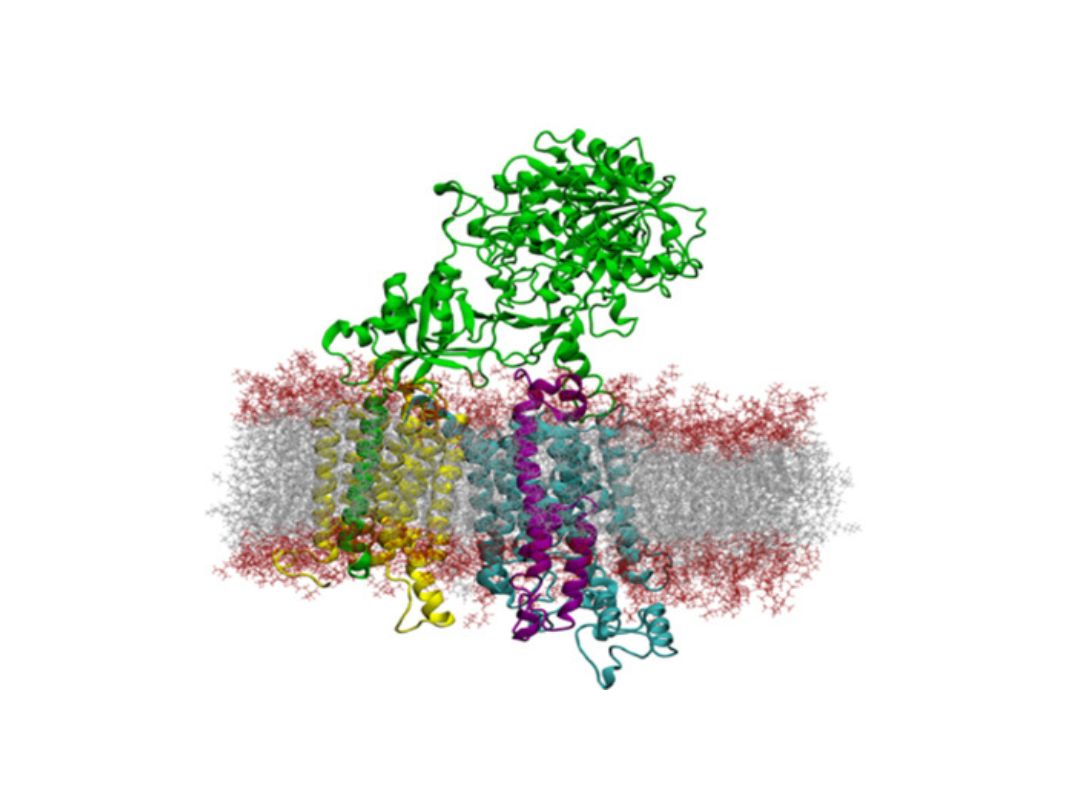
The function of proteins and nucleic acids in living systems is strongly coupled to the molecular motion and dynamics of these biomolecules. We use computer simulation methods to study the structure, function and dynamics of biomolecules. Our goal is to better understand structure formation processes and to elucidate the mechanism of ligand-receptor association in atomic detail.
As the main computational technique we employ Molecular Dynamics simulations based on a classical force field to follow molecular motions including the surrounding solvent and ions. This allows us to extract thermodynamic and kinetic properties of the biomolecular system using methods of statistical mechanics. We also develop computational docking approaches to predict how proteins interact with other proteins or RNA and DNA molecules or how small drug-like compounds bind to biomolecular targets.