Welcome to the Simmel lab - Physics of Synthetic Biological Systems
Our goal is the realization of self-organizing molecular and cellular systems that are able to respond to their environment, compute, move, take action. On the long term, we envision autonomous systems that are reconfigurable, that can evolve and develop.

Single molecule strand displacement: Toehold-mediated strand displacement is at the core of many of the systems developed in dynamic DNA and RNA nanotechnology. The details of this process, in which several nucleic acid molecules with the same or similar sequence compete for binding to a complementary strand, are not fully understood yet. In this work, in collaboration with the groups of Matthias Rief and Petr Šulc we explore the dynamics of strand displacement processes at the single-molecule level using single-molecule force spectroscopy with a optical trap supported by state-of-the-art coarse-grained simulations. Our results reveal the importance of sequence effects for the TMSD process, which is significant for many applications in nucleic acid nanotechnology and synthetic biology.
Walbrun, A., Wang, T., Matthies, M., Šulc, P., Simmel, F. C. & Rief, M. Single-molecule force spectroscopy of toehold-mediated strand displacement. Nat. Commun. 15, 7564 (2024).

Colloidal microswimmers: We explore the use of an electrokinetic phenomenon termed concentration polarization electro-osmosis (CPEO) as a propulsion mechanism for "robotic" microswimmers. CPEO generically occurs when charged colloids in aqueous suspension are subjected to AC electric fields. In this work we created asymmetric particle dimers by linking differently sized colloidal particles with DNA. Depending on the buffer conditions, the particles swim in the direction defined by the larger or smaller particle, driven by CPEO flow around the particle dimer. In contrast to most other propulsion mechanisms demonstrated so far, the 2D movement of the microswimmers can be easily controlled, which we demonstrate by steering the particles with the joystick of a gamepad. We also demonstrate other robotic functions such as pick-up, transport and release of other colloidal particles. We furthermore show that the phenomenon is quite generic and applies to all asymmetric dielectric particles carrying a surface charge, including, e.g., yeast cells.
F. Katzmeier & F. C. Simmel, Microrobots powered by concentration polarization electrophoresis (CPEP). Nature Commun. 14, 6247 (2023).

A nanorobotic wind up toy: Using electrical actuation of a DNA-based nanorobotic arm attached to a base plate via two single-stranded DNA connectors, we show that winding of these strands around each other effectively constitutes a nanoscale molecular torsion spring. Using single-molecule fluorescence tracking, we thoroughly characterize the balance between electrical and mechanical torque for a range of different connectors, and show that such torsion springs can be used to store and also release mechanical energy.
M. Vogt, M. Langecker, M. Gouder, E. Kopperger, F. Rothfischer, F. C. Simmel#, J. List#, Storage of mechanical energy in DNA nanorobotics using molecular torsion springs, Nature Physics 1–11 (2023). doi:10.1038/s41567-023-01938-3

A Brownian ratchet rotor made from DNA origami: In collaboration with the Dietz group, the first Brownian ratchet based on a DNA origami structure was realized, which displays directional rotational movement due to its intrinsic asymmetry when driven out of thermal equilibrium. The structure consists of an arm sitting on a pedestal with three obstacles, leading to six preferred orientations of the arm. When the structure is actuated with an alternating linear electric field (that is not rotating!), the arm rotates directionally. The behavior of the arm is consistent with a "flashing Brownian ratchet" model. Theoretical support for the investigation of the system was provided by the Golestanian group (MPI DS Göttingen).
A.-K. Pumm, W. Engelen, E. Kopperger, J. Isensee, M. Vogt, V. Kozina, M. Kube, M. N. Honemann, E. Bertosin, M. Langecker, R. Golestanian#, F. C. Simmel#, H. Dietz#, A DNA origami rotary ratchet motor, Nature 607, 492–498 (2022). https://doi.org/10.1038/s41586-022-04910-y

Positional information in synthetic cell assemblies: In biological development, an initially homogeneous “material” differentiates into functionally distinct spatial regions, guided by chemical cues from its environment. How well and how robustly a biological system can infer spatial position from the local concentrations of chemicals can be quantified using the theoretical concept of “positional information”. In our work we applied this concept to synthetic cell assemblies. To this end, we created assemblies containing cell-free gene circuits that responded to morphogen gradients in a position-dependent manner. We found that the systems can differentiate into 2-3 regions, and our theoretical analysis allowed us to investigate the parameters that determine and limit the capability of the system to differentiate. We anticipate that similar analyses will be crucial for the further development of autonomous “life-like” materials composed of synthetic cells.
A. Dupin*, L. Aufinger*, I. Styazhkin, F. Rothfischer, B. Kaufmann, S. Schwarz, N. Galensowske, H. Clausen-Schaumann, and F. C. Simmel, Synthetic cell-based materials extract positional information from morphogen gradients, Sci. Adv. 8, eabl9228 (2022).

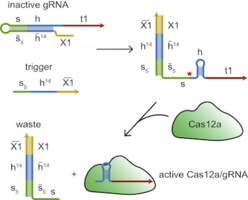
Conditional guide RNAs for Cas12a in mammalian cells: Conditional guide RNAs (gRNAs) allow to make CRISPR-based processes such as gene editing or gene regulation dependent on cellular or environmental signals. We have developed a novel strategy to switch gRNAs for the CRISPR-associated protein Cas12a based on various molecular inputs - microRNAs, short hairpin RNAs, metabolites (via a ribozyme) or other RNA inputs (via a strand displacement process) in mammalian cells. Importantly, in this approach the guide RNAs are produced via a Pol II-promoter and later processed to become a fully functional gRNA. This also allows to encode a full gRNA circuit - including the mRNA encoding the Cas12a - on a single transcript.
L. Oesinghaus and F.C. Simmel, Controlling Gene Expression in Mammalian Cells Using Multiplexed Conditional Guide RNAs for Cas12a, Angew. Chem. Int. Ed. (2021). https://doi.org/10.1002/anie.202107258

Multi-aptamer scaffolds: Aptamers provide a “natural” interface between the DNA and protein world, and have been previously used to bind and arrange proteins on DNA nanostructures. Multivalent binding of target proteins by several aptamers is known to improve the binding strength, but the full potential of origami nanostructures to control the orientation of multiple binders, and also to adjust the flexibility of the binders to enhance their binding properties, has not been utilized so far. We now demonstrate a multiplexed single molecule assay based on DNA origami cavities to optimize the influence of geometry and molecular mechanical properties in two model protein-aptamer systems. Maybe most interestingly, our work also hints at the possibility to “evolve” DNA origami/aptamer scaffolds in a similar way as the SELEX method is used to evolve aptamers in the first place. As a first step in this direction, we demonstrate affinity selection of good binders from a mixture of origami structures.
A. Aghebat Rafat*, S. Sagredo*, M. Thalhammer & F. C. Simmel, Barcoded DNA origami structures for multiplexed optimization and enrichment of DNA-based protein-binding cavities, Nature Chemistry 12, 852-859 (2020). doi:10.1038/s41557-020-0504-6