Martin Stein
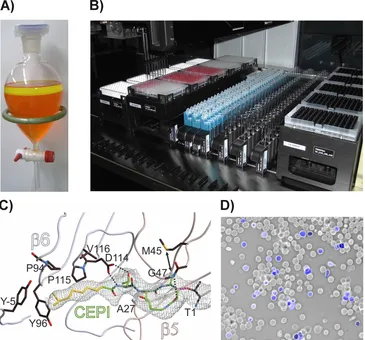
The pivotal role of the eukaryotic proteasome for cell signaling, protein homeostasis and cell cycle control has been proven and discussed in over 25‘000 publications and even more importantly by the admission and successful testing of several anti-cancer and immunosuppressive agents. The latest market entry by Kyprolis(R), which is first natural product based proteasome inhibitor in human application, has revolutionized the approach for the development of further drugs, because its high target specificity has impressively reduced the previously extensive record of side effects. Therefore, the search for other natural lead structures has become an important issue to identify candidates also for applications other than those already addressed by the pharmaceutically applied compounds.However, although high-throughput screening by robot assisted and mass-spec coupled techniques are state-of-the-art today, the search for such extremely toxic substances has to cope with several pitfals. First, most readout techniques rely either directly on UV-VIS-based methodologies or indirectly by the application of fluorescence or luminescence assays. Hence,they suffer from severe background signals due to the high concentration of pigments and other quenching components that can be found abundanatly in natural extracts and fermentation broths (Figure A). Second, micro- and higher organisms tend to restrict the production of toxins to acute defence situations or pathogenic phases in order to avoid self intoxication. For this purpose, they rely on external signals, which induce the biosynthesis of these bioactive compounds to well-defined intervals. Consequently, they are often not comprised in the growth media of conventionally grown cultures or natural extract libraries alike.
In this thesis, both issues were addressed and solved by the introduction of a novel NMR-based peptide digestion assay. In contrast to all previously applied techniques, it is based on a natural proteasomal substrate, that was labelled with a 13C-probe at the scissile peptide bond. Cleavage of the amide to the resulting carboxylic acid and the primary amine results in a frequence shift in the NMR-spectrum. By this approach, we were able to entirely overcome the signal-to-noise problem and at the same time to supply an easily accessible assay suitable for HTS approaches to the scientific community (Figure B). Furthermore, it can and was utilized to detect conditions under which the proteasome inhibitors are actively produced in fermentation broths. In this case, it served in a bioassay guided process to isolate the inhibitors Cepafungin I and Glidobactin A from Photorhabdus luminescens, a close relative to the human pathogens within the Burkholderia genera (Figure C). These inhibitors were subsequently thoroughly characterized by biochemical, biophysical and cell biological means. Hereby, Cepafungin I turned out to be one of the strongest proteasome inhibitors ever analyzed with remarkable biological activity (Figure D). This emphasizes the significance of the technique established in this thesis for future research activity and will contribute to the discovery of other promising compounds.
Awards
2014: VAA Stiftungspreis für pharmazeutische Chemie for the “Detection, identification and characterization of natural 20S proteasome inhibitors form Photorhabdus luminescence”, Berlin, Germany (3,000 €)
2014: Springer Thesis Award Recognizing Outstanding Ph.D. research: "Detection, Identification and Characterization of the Natural 20S Proteasome Inhibitors form Photorhabdus Luminescence" (500 €)
Publications
Pahl A., Lakemeyer M., Vielberg M.-T., Hackl M. W., Vomacka J., Korotkov V. S., Stein M. L., Fetzer C., Lorenz-Baath K., Richter K., Waldmann H., Groll M., Sieber S. A.
Reversible Inhibitors Arrest ClpP in a Defined Conformational State that Can Be Revoked by ClpX Association
Angew. Chem. Int. Ed., 2015, 54, 15892-6, PDF
Fleckenstein T., Kastenmüller A., Stein M. L., Peters C., Daake M., Krause M., Weinfurtner D., Haslbeck M., Weinkauf S., Groll M. and Buchner J.
The Chaperone Activity of the Developmental Small Heat Shock Protein Sip1 Is Regulated by pH-Dependent Conformational Changes
Mol. Cell, 2015, 58, 1067–78, PDF
Voss C., Scholz C., Knorr S., Beck P., Stein M. L., Zall A., Kuckelkorn U., Kloetzel P.-M., Groll M., Hamacher K. and Schmidt B.
α-Keto Phenylamides as P1'-Extended Proteasome Inhibitors
ChemMedChem., 2014, 9, 2557-64, PDF
Trivella D. B., Pereira A., Stein M. L., Byrum T., Valeriote F., Groll M., Gerwick W. H., Morre B. S.
Enzyme inhibition by hydroamination: design and mechanism of a hybrid using a carmaphysin-syringolin enone proteasome inhibitor
Chem. & Biol., 2014, 21, 782-91, PDF
Stein M. L., Cui H., Beck P., Dubiella C., Voss C., Krüger A., Schmidt B., Groll M.
Systematic Comparison of Peptidic Proteasome Inhibitors Highlights the α-Ketoamide Electrophile as an Auspicious Reversible Lead Motif
Angew. Chem. Int. Ed., 2014, 53, 1679-83, PDF
Stein M. L., Groll, M.
Applied techniques for mining natural proteasome inhibitors
Biochim. Biophys. Acta., 2014, 1843, 26-38, PDF
Stein M. L., Beck P., Kaiser M., Dudler R., Becker C. F., Groll M.
One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor
Proc. Natl. Acad. Sci. USA, 2012, 109, 18367-71, PDF
Archer C. R., Groll M., Stein M. L., Schellenberg B., Clerc J., Kaiser M., Kondratyuk T. P., Pezzuto J. M., Dudler R., Bachmann A. S.
Activity Enhancement of Synthetic Syrbactin Proteasome Inhibitor Hybrid and Biological Evaluation in Tumor Cells
Biochemistry, 2012, 51, 6880-8, PDF
Gräwert M. A., Gallastegui N., Stein M. L., Schmidt B., Kloetzel P.-M., Huber R., Groll M.
Elucidation of α-Keto-Aldehyde Binding Mechanism Reveals a Novel Lead Structure Motif for Proteasome Inhibition
Angew. Chem. Int. Ed., 2011, 50, 542-4, PDF