Thomas Badmann
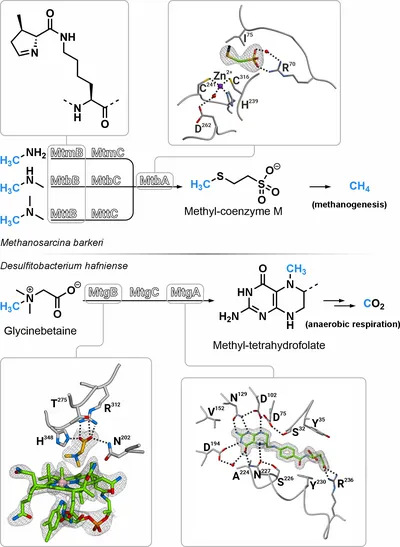
Pyrrolysine is the 22nd proteinogenic amino acid, which is incorporated into proteins in response to amber codons (TAG) in a subset of methanogenic archaea and some bacteria. Only three known enzymes in nature use pyrrolysine for catalysis: the methyltransferases MtmB, MtbB, and MttB, which demethylate their substrates monomethylamine, dimethylamine, and trimethylamine, respectively, providing methyl groups for energy production via methanogenesis. Interestingly, homologous proteins of the methyltransferase MttB without pyrrolysine are widely distributed. These include MtgB, MtpB, MtcB, and MtyB, which have been identified as demethylases of various quaternary amines. However, structural details about their mode of action are lacking from literature. Carrier proteins with the cofactor cobalamin coordinate the methyl cleavage from methylated amines and transfer the methyl group to associated methyltransferases that catalyze the methylation of cofactors such as coenzyme M or tetrahydrofolate. The proteins involved in cobalamin-mediated methyl transfer show a high degree of structural similarity, even in distantly related organisms. Therefore, insights into the function of one enzyme could potentially be applied to a large number of related proteins, helping to elucidate the still not fully understood mechanism of cobalamin-mediated methyl transfer.
In my PhD thesis I solved several methyltransferase structures of the microbial methylamine metabolism using X‑ray crystallography. The active site of MtgB, a pyrrolysine-deficient homolog of the MttB family, shows that the absence of this catalytic amino acid is compensated by a fundamentally different amino acid composition of the active pocket. Substrate- and cobalamin-bound structures of MtgB show how glycinebetaine is positioned for methyl transfer to cobalamin. Based on a substrate and a product structure of the zinc-dependent coenzyme M methylase MtbA, an inversion of the tetrahedral metal coordination can be observed during the reaction of this enzyme. Upon methylation, coenzyme M is lost as an interaction partner of the zinc ion and is replaced by an aspartate on the opposite side. Substrate- and product-bound structures of MtgA, combined with activity measurements, were used to characterize this methyltransferase in detail. MtgA locks its cofactor tetrahydrofolate after methylation in a novel way, thus stabilizing the protonated intermediate reaction state. The information obtained from the structures contributes to fundamental research in the fields of methanogenesis, the cofactors cobalamin, coenzyme M and tetrahydrofolate, and pyrrolysine-mediated methyl transfer.


Publications
Mordhorst S., Badmann T., Bösch N. M., Morinaka B., Rauch H., Piel J., Groll M., Vagstad A. L.
Structural and Biochemical Insights into Post-Translational Arginine-to-Ornithine Peptide Modifications by an Atypical Arginase
ACS Chem. Biol., 2023, 18, 528-36
Badmann T., Groll M.
Atomic resolution structures of tetrahydrofolate methylation in the desulfitobacterial glycine betaine metabolism.
ChemBioChem., 2020, 21, 776-9