Marie-Theres Vielberg
Proteolysis is not only essential for energy production and recycling of amino acids to synthesize new proteins but also for degradation of catalytically impaired or no longer required proteins. Since bacteria possess no specialized organelles to undertake these tasks, they rely on a huge variety of proteases. Two of them have been the research focus during my PhD studies: i) the ‘ancestral β-subunit protein’ (Anbu) and ii) the caseinolytic protease P (ClpP).
To be or not to be (a protease): Anbu
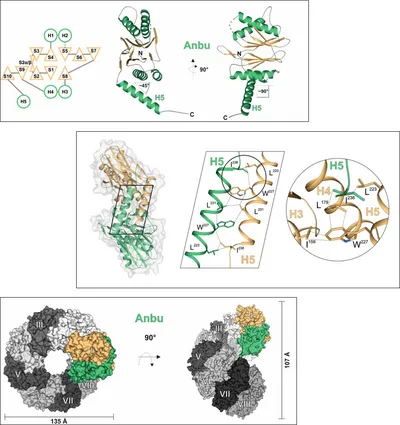
The 20S proteasome, a multi-subunit protease, performs most of non-lysosomal proteolysis in eukaryotes. It is also ubiquitously present in archaea, while its occurrence in bacteria is rare. Many of these organisms encode the homologous protease HslV instead. However, having a huge number of species, including all cyanobacteria, encoding neither the 20S proteasome nor HslV, the evolutionary origin of this family of N-terminal threonine proteases was questioned. A recent study, searching for further bacterial progenitors of the 20S proteasome, discovered the existence of another member of this protease family: the ‘ancestral β-subunit protein’ (Anbu). Studying the Anbu protein from Hyphomicrobium sp. strain MC1 resulted in growth of well-diffracting crystals, allowing us to solve its structure via SAD-phasing. Although it seems to assemble in defined particles in solution, Anbu formed helical superstructures in the crystal, contrary to the closed, barrel-shaped structures known from the 20S proteasome or HslV. Nevertheless, the fold of an individual subunit clearly identifies Anbu as an Ntn-hydrolase and the inter-subunit contacts within the structures of all three protein complexes are similar. Conservation of the Thr1Oγ activating triad suggested that Anbu could also function as an endoprotease. However, using standard proteasomal substrates no such activity was detected. Potential reasons might be the missing activation of the Thr1-NH2 terminus, the lack of a suitable substrate or a missing interaction partner. To get a deeper understanding of this presumed protease, we also teamed up with the group of Prof. Matthias Bochtler (IIMCB, Warsaw), which investigated the Anbu protein from Yersinia bercovieri. These studies showed Anbu to form ‘lock-washer-shaped’ particles with resemblances to the structures of the 20S proteasome and HslV as well as to the helical superstructures seen before, but even there no proteolytic activity was detected. So, the question remains whether the evolutionary relationship between Anbu, the 20S proteasome and HslV is purely structural or also a functional one.
Targeting antibiotic resistance: ClpP
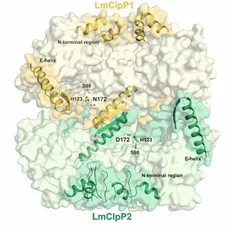
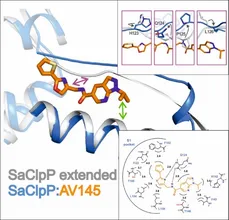
The caseinolytic protease P (ClpP) is essential for the virulence of several facultative and obligate pathogens. Since there is a rapid increase in antibiotic resistances over the last decades, ClpP has been listed as a potential candidate for drug development. The research on ClpP took part in collaboration with the group of Prof. Stephan Sieber (TUM, Munich). In contrast to most bacteria, Listeria monocytogenes encodes two isoforms of ClpP. Although the sequence identity of the two proteins amounts to less than 45 %, they can form a heterocomplex together. We used X-ray crystallographic analysis to understand how it differs from homomeric ClpPs. The data reveal that the overall architecture and stabilization of homo- and heteromeric ClpP particles adopt the same topology. However, there exist differences in the axial pore formation, active site composition and shape of the S1 pocket between ClpP1 and ClpP2. These results suggest a potential task sharing in the heterocomplex and provide valuable information for the design of isoform-specific inhibitors to characterise the function of the ClpP1/2 heterocomplex in more detail. Despite the high potential of ClpP as new antibiotic target, the number of specific inhibitors described so far is limited. A high-throughput screen with about 140,000 compounds identified the first non-covalently inhibitor AV145 of ClpP from Staphylococcus aureus. Co-crystallization allowed us to reveal an unexpected binding mode of the compound inducing a novel, inactive conformation of the protease. Squeezed between α-helix E and β-sheet 9, the inhibitor causes severe distortions in these two secondary structures. Rotation of Pro125 and the resulting misalignment of the catalytic triad particularly explain the special mode of action. However, in vivo experiments showed that binding of ClpP-specific regulators overwrites the inhibitory effect of AV145, pointing out the challenges in drug development for ClpP.
Publications
Vogler M., Karan R., Renn D., Vancea A., Vielberg M.-T., Grötzinger S. W., DasSarma P., DasSarma S., Eppinger J., Groll M., Rueping M.
Crystal Structure and Active Site Engineering of a Halophilic γ-Carbonic Anhydrase
Front. Microbiol., 2020, 11, 1-12, PDF
Malik I. T., Pereira R., Vielberg M.-T., Mayer C., Straetener J., Thomy D., Famulla K., Castro H., Sass P., Groll M., Brötz-Oesterhelt H.
Functional characterisation of ClpP mutations conferring resistance to acyldepsipeptide antibiotics in Firmicutes
ChemBioChem., 2020, 21, 1997-2012, PDF
Kazman P., Vielberg M.-T., Pulido Cendales M. D., Hunziger L., Weber B., Hegenbart U., Zacharias M., Köhler R., Schönland S., Groll M., Buchner, J.
A patient-derived antibody light chain shows fatal amyloid formation caused by a single point mutation
Elife, 2020, 9 (e52300), 1-23, PDF
Vielberg M.-T., Bauer V. C., Groll M.
On the Trails of the Proteasome Fold: Structural and Functional Analysis of the Ancestral β-Subunit Protein Anbu
J. Mol. Biol., 2018, 430, 628-40, PDF
Piasecka A., Czapinska H., Vielberg M.-T., Szczepanowski R., Kiefersauer R-, Reed S., Groll M., Bochtler M.
The Y. bercovieri Anbu crystal structure sheds light on the evolution of highly (pseudo)symmetric multimers
J. Mol. Biol., 2018, 430, 611-27, PDF
Annamalai K., Liberta F., Vielberg M.-T., Close W., Lilie H., Gührs K. Hl, Schierhorn A., Koehler R., Schmidt A., Haupt C., Hegenbart U., Schönland S., Schmidt M, Groll M., Fändrich M.
Common Fibril Structures Imply Systemically Conserved Protein Misfolding Pathways In Vivo
Angew. Chem. Int. Ed., 2017, 56, 7510-4, PDF
Pahl A., Lakemeyer M., Vielberg M.-T., Hackl M. W., Vomacka J., Korotkov V. S., Stein M. L., Fetzer C., Lorenz-Baath K., Richter K., Waldmann H., Groll M., Sieber S. A.
Reversible Inhibitors Arrest ClpP in a Defined Conformational State that Can Be Revoked by ClpX Association
Angew. Chem. Int. Ed., 2015, 54, 15892-6, PDF
Dahmen M., Vielberg M.-T., Groll M., Sieber S. A.
Structure and mechanism of the caseinolytic protease ClpP1/2 heterocomplex from Listeria monocytogenes
Angew. Chem. Int. Ed., 2015, 54, 3598-602, PDF