Felix Quitterer
The 22nd proteinogenic amino acid pyrrolysine is incorporated into methylamine methyltransferases of methane-producing Methanosarcinaceae and some bacteria via read-through of an amber stop codon (UAG). This requires the presence of a specific aminoacyl-tRNA synthetase (PylS) that links pyrrolysine to its cognate tRNA (tRNAPyl), which comprises a CUA anticodon. The novel amino acid acts as the catalytic residue within these methylamine methyltransferases and initiates the breakdown of methylamines to methane in the methanogenic pathway. Recent studies revealed L-lysine as the sole precursor for pyrrolysine. The enzymes PylB, PylC, and PylD were shown to catalyze the conversion from L-lysine to pyrrolysine, but their exact function remained unclear.
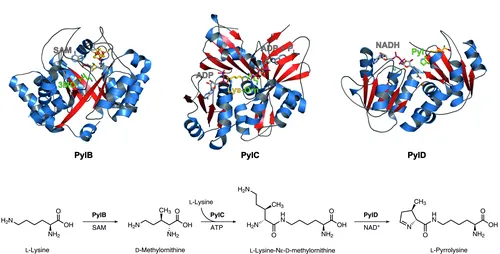
During my PhD thesis I determined the crystal structures of PylB, PylC, and PylD in complex with their substrates, intermediates or products. The molecular architecture of the enzymes as well as the location and orientation of cofactors and ligands within their active sites allowed the assessment of detailed reaction trajectories for all three enzymes studied.
PylB, a member of the radical S-adenosyl-L-methionine (SAM) family with a classical (β/α)8 TIM-barrel fold, uses a [4Fe-4S] cluster and SAM to initiate a fragmentation-recombination sequence affording 3R-methyl-D-ornithine from L-lysine.
PylC then catalyzes the ATP-dependent condensation of 3R-methyl-D-ornithine and a second lysine molecule to L-lysine-Nε-3R-methyl-D-ornithine. As deduced from the crystallographic results, catalysis requires ordered substrate entry to the active site cavity, i. e. methylornithine has to bind prior to lysine. Moreover, and different from other ATP-grasp enzymes, PylC shows a second ATP-binding site. The additional nucleotide is not required for phosphorylation of the substrate, but, as revealed by mutagenic studies, is necessary for coordination and orientation of L-lysine and the correct folding of the protein.
PylD, the final enzyme of the pyrrolysine biosynthesis, exhibits an induced-fit mechanism upon substrate binding. Structural and kinetic characterization of the dehydrogenase displayed a tolerance for different substrate analogs that can be oxidized and cyclized using NAD+ as coenzyme. Whereas other dehydrogenases possess a base within the active site cavity that facilitates proton abstraction from the substrate, there is no amino residue at the PylD active site that could support proton exchange reactions. However, a well-defined water cluster in close proximity to the ornithine moiety of L-lysine-Nε-3R-methyl-D-ornithine may be able to carry out this function.
My achieved structural and functional results of the pyrrolysine biosynthesis provide a path for using PylB, PylC, and PylD to generate pyrrolysine analogs in vivo. In the future, random mutagenesis of theses enzymes, as already demonstrated for PylS, might lead to an expanded substrate acceptance. A system consisting of the pylBCD gene cluster in combination with pylTS might be a promising route to directly biosynthesize unnatural amino acids and incorporate them into target proteins in order to use their specific properties for protein engineering.
Publications
Fischer J., Renn D., Quitterer F., Radhakrishnan A., Liu M., Makki A., Ghorpade S., Rueping M., Arold S. T., Groll M., Eppinger J.
A Robust and Versatile Host Protein for the Design and Evaluation of Artificial Metal Centers
ACS Catal., 2019, 9, 11371−80, PDF
Quitterer F., Frank A., Wang K., Rao G., O'Dowd B., Li J., Guerra F., Abdel-Azeim S., Bacher A., Eppinger J., Oldfield E., Groll M.
Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE)
J. Mol. Biol., 2015, 427 (12), 2220-8, PDF
Quitterer F., Beck P., Bacher A., Groll M.
The formation of Pyrroline and tetrahydropyridine rings in amino acids catalyzed by pyrrolysine synthase (PylD)
Angew. Chem. Int. Ed., 2014, 53, 8150-3, PDF
Weiz A. R., Ishida K., Quitterer F., Meyer S., Kehr J., Müller K., Groll M., Hertweck C., Dittmann E.
Harnessing the Evolvability of Tricyclic Microviridins To Dissect Protease–Inhibitor Interactions
Angew. Chem. Int. Ed., 2014, 53, 3735-8, PDF
Quitterer F., Beck P., Bacher A., Groll M.
Structure and reaction mechanism of pyrrolysine synthase (PylD)
Angew. Chem. Int. Ed., 2013, 52, 7033-7, PDF
Quitterer F., List A., Beck P., Bacher A., Groll M.
Biosynthesis of the 22nd Genetically Encoded Amino Acid Pyrrolysine: Structure and Reaction Mechanism of PylC at 1.5 Å Resolution
J. Mol. Biol., 2012, 424, 270-82, PDF
Quitterer F., List A., Eisenreich W., Bacher A., Groll M.
Crystal Structure of Methylornithine Synthase (PylB): Insights into the Pyrrolysine Biosynthesis
Angew. Chem. Int. Ed., 2012, 51, 1339-42, PDF
Lee M., Gräwert T., Quitterer F., Rohdich F., Eppinger J., Eisenreich W., Bacher A., Groll M.
Biosynthesis of isoprenoids: crystal structure of the [4Fe-4S] cluster protein IspG
J. Mol. Biol., 2010, 404, 600-10, PDF