Anja List
The key component of the ubiquitin-mediated degradation pathway, the 20S proteasome, maintains biological homeostasis and regulates many crucial processes in the cell by cleaving endoproteolytic most intracellular proteins. This complex hydrolysis machinery has received considerable attention for the treatment of many diseases, including cancer. The role of proteasome:ligand complexes is well established and has been shown to be an important tool in inhibitor analysis of this crucial protein degradation machinery. However their molecular binding mode requires additional investigation because of the ineffectiveness of current market drugs against some types of solid tumors.
We solved a new crystal structure of a natural product in complex with the proteasome, that can be used for inhibitor optimization using both crystallographic and cell toxicity tests. We found that for proteasome inhibition the whole architecture is essential for selectivity and that more than one functional β-lactone group is necessary, which can be explained exactly by the crystal structure. Furthermore, we performed cell toxicity assays and target identification using click chemistry and mass spectrometry. Our results demonstrate that similar structures of β-lacton-natural products exist in bacteria and fungi, but they have different target specificity because of their biological background. We anticipate our results to be a starting point for understanding of the β-lactone based inhibitor binding mechanism to the eukaryotic CP. Furthermore, the complexed crystal structure provided a valuable explanation for the unreactivity of proteasome ligands against the 20S eukaryotic proteasome. The proteasome inhibition is still a major target of anti-cancer drug development, and a well-defined structural analysis of the binding mode will be relevant for such developments.
Thesis
A. List (2012), Biochemische und strukturelle Charakterisierung von Proteasen und ihren Liganden
Publications
List A., Zeiler E., Gallastegui N., Rusch M, Hedberg C, Sieber S. A., Groll M.
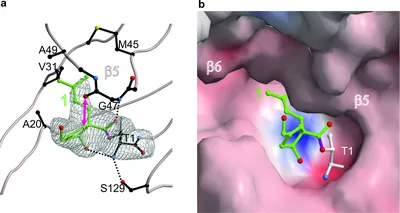
Omuralide and Vibralactone: Differences in the Proteasome- β-Lactone-γ-Lactam Binding Scaffold Alter Target Preferences
Angew. Chem. Int. Ed., 2014, 53, 571-4, PDF
Zeiler E., List A., Alte F., Gersch M., Wachtel R., Poreba M., Drag M., Groll M., Sieber S. A.
Structural and functional insights into caseinolytic proteases reveal an unprecedented regulation principle of their catalytic triad
Proc. Natl. Acad. Sci. USA, 2013, 110, 11302-7, PDF
Kawamura S., Unno Y., List A., Mizuno A., Tanaka M., Sasaki T., Arisawa M., Asai A., Groll M., Shuto S.
Potent proteasome inhibitors derived from the unnatural cis-cyclopropane isomer of Belactosin A: synthesis, biological activity, and mode of action
J. Med. Chem., 2013, 56, 3689-700, PDF
Quitterer F., List A., Beck P., Bacher A., Groll M.
Biosynthesis of the 22nd Genetically Encoded Amino Acid Pyrrolysine: Structure and Reaction Mechanism of PylC at 1.5 Å Resolution
J. Mol. Biol., 2012, 424, 270-82, PDF
Gersch M., List A., Groll M., Sieber S. A.
Insights into the structural network responsible for oligomerization and activity of the bacterial virulence regulator caseinolytic protease P (ClpP)
J. Biol. Chem., 2012, 287, 9484-94, PDF
Quitterer F., List A., Eisenreich W., Bacher A., Groll M.
Crystal Structure of Methylornithine Synthase (PylB): Insights into the Pyrrolysine Biosynthesis
Angew. Chem. Int. Ed., 2012, 51, 1339-42, PDF